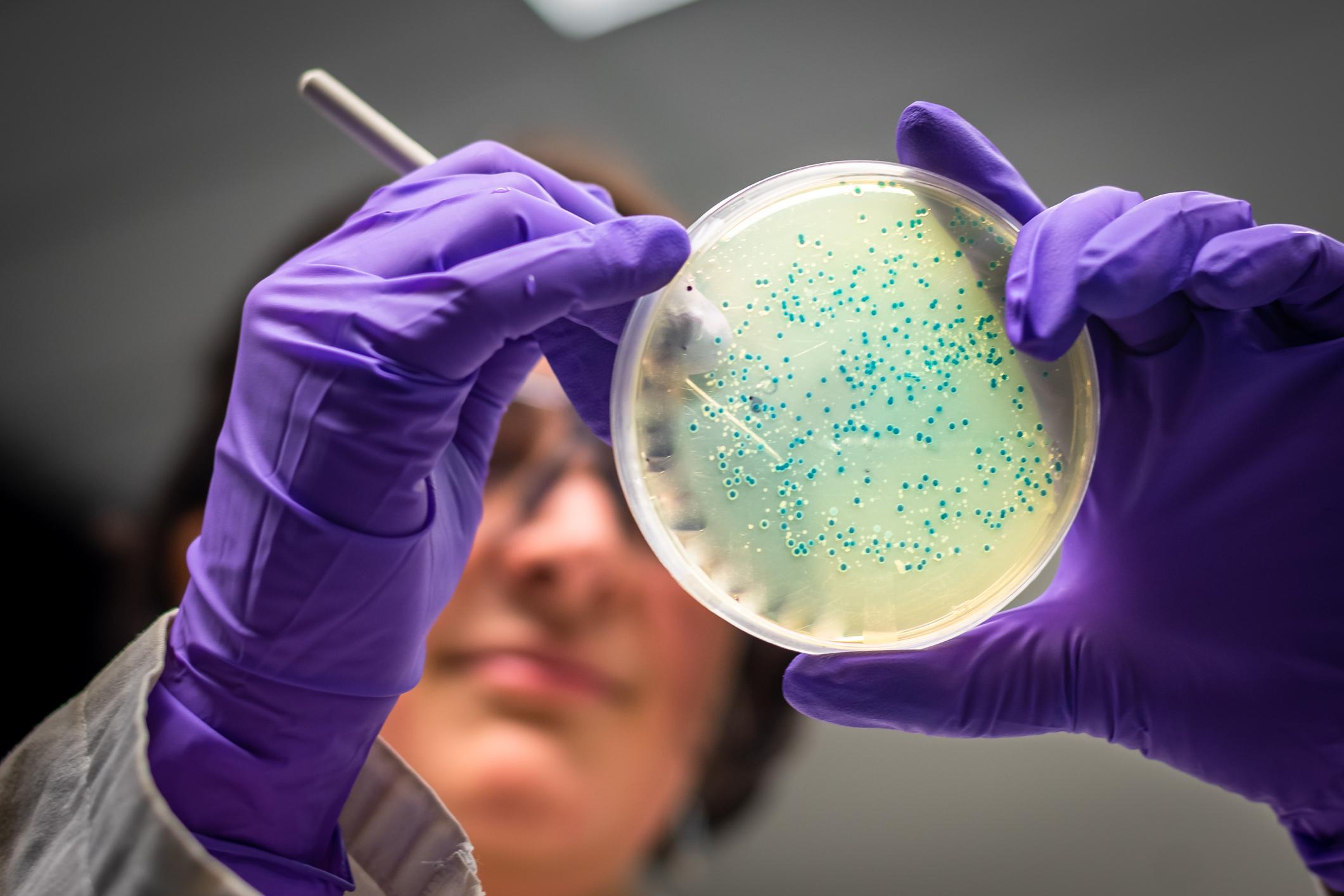
Viruses and bacteria are capable of exchanging their antibiotic resistant properties with an almost unlimited amount of their fellows. In addition, in contact with a new ecosystem, they are able to adapt to protect themselves, and to share their knowledge with others, which explains an increasingly strong resistance to antibiotics.
![]()
- Viruses and bacteria can exchange parts of their genetic material responsible for antibiotic resistance.
- Their resistance increases when they encounter a new ecosystem, both natural and human.
- The antibiotic resistance of viruses and bacteria can potentially become almost unlimited.
Mapping all living things to protect against them is no easy task. If the company is complex, it aims above all to better protect humans against viruses and bacteria that can attack us, all the more so when we do not know how to get rid of them. Researchers from the Chalmers University of Technology (Sweden) have demonstrated that significant genetic transfers between humans and bacteria in our ecosystem could be the cause of antibiotic resistance. The results of their work have been published on the MicrobiologyOpen platform.
Exchanges between micro-organisms to become more resistant
We speak of antibiotic resistance when microorganisms such as viruses or bacteria have acquired the ability to resist antibiotics. According to the World Health Organization (WHO), antibiotic resistance is one of the greatest threats to health, food security and development worldwide, as it becomes increasingly difficult to get rid of it. potentially pathogenic agents. In Europe alone, antibiotic resistance already causes more than 33,000 deaths a year.
Antibiotic resistance is not specific to a single class of bacteria or virus. Entirely different species of bacteria can transmit resistance genes to each other via plasmids, small DNA molecules in which they store some of their genes outside the chromosome. Thus, when two bacterial cells come into contact, they can mutually copy their plasmids, which makes both species resistant to antibiotics. In this case, we speak of conjugation.
“In recent years, we have seen that resistance genes have spread to human pathogens to a much greater extent than expected.says Jan Zrimec, a systems and synthetic biology researcher at Chalmers University of Technology. Many of these genes appear to come from a wide range of bacterial species and environments, such as soil, water, and plant bacteria. This was difficult to explain, because although conjugation is very common, we thought there was a distinct limit for which bacterial species can transfer plasmids between each other.”
With his team of researchers, Jan Zrimec has developed a method to understand the extent of the phenomenon in bacteria and viruses. Using an algorithm capable of identifying specific regions of DNA necessary for conjugation, the researchers were able to explore the genetic sequences of 4,600 plasmids from several types of bacteria. Clearly, gene transfer could be far more limitless and widespread than they anticipated.
Almost unlimited potential for antibiotic resistance
The oriT region alone, the DNA-specific one for conjugation, is actually eight times larger than the researchers estimate, and harbors twice as many plasmids as predicted. Similarly, the number of bacterial species that possess mobile plasmids could be almost twice as high as previously known. Of these plasmids, more than half have conjugating enzymes that come from another bacterium.
“These results could imply that there is a strong network for the transfer of plasmids between bacteria from humans, animals, plants, soil, aquatic environments and industries, to name a few.analyzes Jan Zrimec. Resistance genes occur naturally in many different bacteria in these ecosystems, and the hypothetical network could mean that genes from all these environments can be transferred to bacteria that cause disease in humans. This could be a possible reason for the rapid development of resistance in human pathogens that we have observed in recent years. Our intensive use of antibiotics selects for resistance genes, which could therefore come from a much larger natural genetic reservoir than previously estimated.”
Further studies should be performed in the future to better understand the scope of these plasmid exchanges. However, the data analysis methods developed by Zrimec can already be used by many researchers working on antibiotic resistance in various medical and biological fields. They constitute a powerful new tool to systematically map the potential transferability of different plasmids.
.